Some miscellaneous tips I've discovered after over a year of being hands-on with a clinical ETL pipeline.
Technology
Set up a local dev environment (Git, Pip, IDE of choice, Python environment etc). Being able to test locally cannot be more important. For instance, I've been using Palantir's Foundry but since it's just a wrapper around Spark, you can have a full, locally run, test suite using PySpark.
If you can't get the data on your laptop (GDPR etc), use simulated data. If your model cannot find a signal in the noise you control, it likely won't when it faces real data. Make these tests automated so it can be part of your CI/CD pipeline.
Keep everything as deterministic as you can. Using commands like SQL's first means you have no guarantees what will come out of the sausage machine. How can your QAs test that?
Prefer to write intermediary data sets to persistent storage and load them again in the next stage over processing everything in memory with a single output. It might make the pipeline take longer to run but it gives you something like an "audit log" of the steps in your model.
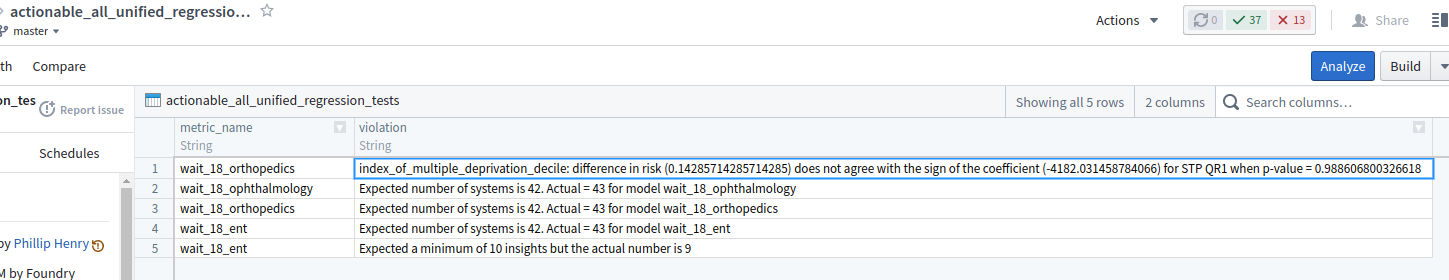
Have diagnositic transforms in your pipeline. Typically, they make assertions to ensure self-consistency (eg, all patients' hospital length-of-stay must be non-negative; the aggregated total of overnight patients cannot exceed the total number of beds). These transforms output any violations into data sets that will never be seen by the end user, just the MLOps team. Diagnostic transforms have the advantage over the local test in that they can use real data as they are embedded in the production pipeline.
Process
Testing data is harder than testing code so spare a thought for your QAs. Talk to the them long before you cut a line of code and establish whether they can test what you're building. If not, find out what you need to give them.
Talk to data owners, tell them what you're trying to do and ask them how they'd do it. They often tell you things you'd never know a priori - like which data is, ahem, "less reliable".
Spend some time (a week?) just playing with the data (EDA) so things become more apparent. It's important to know, for instance, if a column called "region" in A&E data set is the region of the patient or the care provider. Since a patient in A&E may be treated the other side of the country (perhaps they were taken ill while on holiday), the two regions might have nothing to do with one another.
Make sure everybody is singing from the same hymn sheet with a ubiquitous language. Calculating the number of weeks a patient waits couldn't be easier, right? Take the number of days between the start and end date and divide by 7, right? But do we round up or down? And MS SQLServer has its own weird way of doing this calculation.
Data
Don't forget the effect of time on your data (data drift). For instance, care provider networks often get restructured so does hospital trust X in 2020 go by a different name today? Patients move around the country so the same person may be associated with more than one locale. Did more data sources come online over time?
How often is the data updated and how stale is it? These are not the same thing. For instance, the data may be updated daily but its content lags behind by several months.
Were dodgy assumptions made on upstream data sets that you need to be aware of? For instance, removing dead people from the the patients data set might have made sense upstream when dealing with vaccination data but not for historical accident and emergency data.
When unifying data sets, watch out for semantically equivalent values that may be lexically different (eg, if one data set uses an ethnicity of "British white" and another that says "White British", then a union of the two will look like two different ethnicities). This problem becomes harder when one data set uses, say, different age bands. How would you reconcile them?